Abstract
Purpose
Modic changes (MC) on magnetic resonance imaging (MRI) have been associated with the development and severity of low back pain (LBP). The etiology of MC remains elusive, but it has been suggested that altered metabolism may be a risk factor. As such, this study aimed to identify metabolomic biomarkers for MC phenotypes of the lumbar spine via a combined metabolomic-genomic approach.
Methods
A population cohort of 3,584 southern Chinese underwent lumbar spine MRI. Blood samples were genotyped with single-nucleotide polymorphisms (SNP) arrays (n = 2,482) and serum metabolomics profiling using magnetic resonance spectroscopy (n = 757), covering 130 metabolites representing three molecular windows, were assessed. Genome-wide association studies (GWAS) were performed on each metabolite, to construct polygenic scores for predicting metabolite levels in subjects who had GWAS but not metabolomic data. Associations between predicted metabolite levels and MC phenotypes were assessed using linear/logistic regression and least absolute shrinkage and selection operator (LASSO). Two-sample Mendelian randomization analysis tested for causal relationships between metabolic biomarkers and MC.
Results
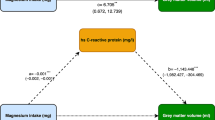
20.4% had MC (10.6% type 1, 67.2% type 2, 22.2% mixed types). Significant MC metabolomic biomarkers were mean diameter of very-low-density lipoprotein (VLDL)/low-density lipoprotein (LDL) particles and cholesterol esters/phospholipids in large LDL. Mendelian randomization indicated that decreased VLDL mean diameter may lead to MC.
Conclusions
This large-scale study is the first to address metabolomics in subject with/without lumbar MC. Causality studies implicate VLDL related to MC, noting a metabolic etiology. Our study substantiates the field of “spino-metabolomics” and illustrates the power of integrating metabolomics-genomics-imaging phenotypes to discover biomarkers for spinal disorders, paving the way for more personalized spine care for patients.





Similar content being viewed by others
References
Coste J, Paolaggi JB, Spira A (1991) Reliability of interpretation of plain lumbar spine radiographs in benign, mechanical low-back pain. Spine 16(4):426–428 (PubMed PMID: 1828626)
de Schepper EI, Damen J, van Meurs JB, et al. (2010) The association between lumbar disc degeneration and low back pain: the influence of age, gender, and individual radiographic features. Spine (Phila Pa 1976). 35(5): 531–6. Epub 2010/02/12. doi: https://doi.org/10.1097/BRS.0b013e3181aa5b33. PubMed PMID: 20147869.
Luoma K, Riihimaki H, Luukkonen R et al (2000) Low back pain in relation to lumbar disc degeneration. Spine 25(4):487–492 (PubMed PMID: 10707396)
Maatta JH, Karppinen J, Paananen M, et al. (2016) Refined Phenotyping of Modic Changes: Imaging Biomarkers of Prolonged Severe Low Back Pain and Disability. Medicine (Baltimore). 95(22): e3495. doi: https://doi.org/10.1097/MD.0000000000003495. PubMed PMID: 27258491; PubMed Central PMCID: PMCPMC4900699.
Maatta JH, Karppinen JI, Luk KD et al (2015) Phenotype profiling of Modic changes of the lumbar spine and its association with other MRI phenotypes: a large-scale population-based study. Spine J 15(9):1933–1942. https://doi.org/10.1016/j.spinee.2015.06.056 (PubMed PMID: 26133258)
Samartzis D, Mok FPS, Karppinen J et al (2016) Classification of Schmorl’s nodes of the lumbar spine and association with disc degeneration: a large-scale population-based MRI study. Osteoarthritis Cartilage 24(10):1753–1760. https://doi.org/10.1016/j.joca.2016.04.020 (PubMed PMID: 27143364)
Battie MC, Videman T, Gill K, et al. (1991) 1991 Volvo Award in clinical sciences. Smoking and lumbar intervertebral disc degeneration: an MRI study of identical twins. Spine. 16(9): 1015–21. PubMed PMID: 1948392.
Eskola PJ, Lemmela S, Kjaer P et al (2012) Genetic association studies in lumbar disc degeneration: a systematic review. PLoS ONE 7(11):e49995. https://doi.org/10.1371/journal.pone.0049995.PubMedPMID:23185509;PubMedCentralPMCID:PMCPMC3503778
Kauppila LI. (2009) Atherosclerosis and disc degeneration/low-back pain--a systematic review. Eur J Vasc Endovasc Surg.37(6):661–70. Epub 2009/03/31. doi: S1078–5884(09)00090–2 [pii] https://doi.org/10.1016/j.ejvs.2009.02.006. PubMed PMID: 19328027.
Li Y, Samartzis D, Campbell DD et al (2016) Two subtypes of intervertebral disc degeneration distinguished by large-scale population-based study. Spine J 16(9):1079–1089. https://doi.org/10.1016/j.spinee.2016.04.020 (PubMed PMID: 27157501)
Samartzis D, Karppinen J, Chan D et al (2012) The association of lumbar intervertebral disc degeneration on magnetic resonance imaging with body mass index in overweight and obese adults: a population-based study. Arthritis Rheum 64(5):1488–1496. https://doi.org/10.1002/art.33462 (PubMed PMID: 22287295)
Samartzis D, Karppinen J, Mok F, et al. (2011) A population-based study of juvenile disc degeneration and its association with overweight and obesity, low back pain, and diminished functional status. J Bone Joint Surg Am.93(7):662–70. Epub 2011/04/08. doi: 93/7/662 [pii] https://doi.org/10.2106/JBJS.I.01568. PubMed PMID: 21471420.
Teraguchi M, Yoshimura N, Hashizume H et al (2016) Metabolic Syndrome Components Are Associated with Intervertebral Disc Degeneration: The Wakayama Spine Study. PLoS ONE 11(2):e0147565. https://doi.org/10.1371/journal.pone.0147565.PubMedPMID:26840834;PubMedCentralPMCID:PMCPMC4739731
Ohshima H, Urban JP. (1992) The effect of lactate and pH on proteoglycan and protein synthesis rates in the intervertebral disc. Spine (Phila Pa 1976).17(9):1079–82. Epub 1992/09/01. PubMed PMID: 1411761.
Urban JP, Smith S, Fairbank JC. (2004) Nutrition of the intervertebral disc. Spine (Phila Pa 1976).29(23):2700–9. doi: https://doi.org/10.1097/01.brs.0000146499.97948.52. PubMed PMID: 15564919.
Keshari KR, Lotz JC, Link TM, et al. (2008) Lactic acid and proteoglycans as metabolic markers for discogenic back pain. Spine (Phila Pa 1976).33(3):312–7. Epub 2008/02/28. doi: https://doi.org/10.1097/BRS.0b013e31816201c300007632-200802010-00015 [pii]. PubMed PMID: 18303465.
Cameron BM, VanderPutten DM, Merril CR. (1995) Preliminary study of an increase of a plasma apolipoprotein E variant associated with peripheral nerve damage. A finding in patients with chronic spinal pain. Spine (Phila Pa 1976).20(5):581–9; discussion 9–90. Epub 1995/03/01. PubMed PMID: 7604328.
VanderPutten DM, Cameron BM, Merril CR (1993) Increased apolipoprotein-E concentrations in individuals suffering chronic low back syndrome identified by two-dimensional gel electrophoresis. Appl Theor Electrophor 3(5):247–252 (Epub 1993/01/01 PubMed PMID: 8218479)
Smith VH. (2010) Vitamin C deficiency is an under-diagnosed contributor to degenerative disc disease in the elderly. Med Hypotheses.74(4):695–7. Epub 2009/11/26. doi: S0306–9877(09)00720–8 [pii] https://doi.org/10.1016/j.mehy.2009.10.041. PubMed PMID: 19932568.
Modic MT, Steinberg PM, Ross JS, et al. (1988) Degenerative disk disease: assessment of changes in vertebral body marrow with MR imaging. Radiology.166(1 Pt 1):193–9. Epub 1988/01/01. doi: https://doi.org/10.1148/radiology.166.1.3336678. PubMed PMID: 3336678.
Thompson KJ, Dagher AP, Eckel TS et al (2009) Modic changes on MR images as studied with provocative diskography: clinical relevance–a retrospective study of 2457 disks. Radiology 250(3):849–855. https://doi.org/10.1148/radiol.2503080474 (PubMed PMID: 19244050)
Jensen TS, Karppinen J, Sorensen JS et al (2008) Vertebral endplate signal changes (Modic change): a systematic literature review of prevalence and association with non-specific low back pain. Eur Spine J 17(11):1407–1422. https://doi.org/10.1007/s00586-008-0770-2.PubMedPMID:18787845;PubMedCentralPMCID:PMCPMC2583186
Maatta JH, Wadge S, MacGregor A, et al. (2015) ISSLS Prize Winner: Vertebral Endplate (Modic) Change is an Independent Risk Factor for Episodes of Severe and Disabling Low Back Pain. Spine (Phila Pa 1976).40(15):1187–93. doi: https://doi.org/10.1097/BRS.0000000000000937. PubMed PMID: 25893353.
Mok FP, Samartzis D, Karppinen J et al (2016) Modic changes of the lumbar spine: prevalence, risk factors, and association with disc degeneration and low back pain in a large-scale population-based cohort. Spine J 16(1):32–41. https://doi.org/10.1016/j.spinee.2015.09.060 (PubMed PMID: 26456851)
Takatalo J, Karppinen J, Niinimaki J, et al. (2012) Association of modic changes, Schmorl's nodes, spondylolytic defects, high-intensity zone lesions, disc herniations, and radial tears with low back symptom severity among young Finnish adults. Spine (Phila Pa 1976).37(14):1231–9. doi: https://doi.org/10.1097/BRS.0b013e3182443855. PubMed PMID: 22166927.
Freidin M, Kraatari M, Skarp S et al (2019) Genome-wide meta-analysis identifies genetic locus on chromosome 9 associated with Modic changes. J Med Genet 56(7):420–426. https://doi.org/10.1136/jmedgenet-2018-105726 (PubMed PMID: 30808802)
Wu J, Huang J, Battie MC et al (2020) Lifestyle and lifetime occupational exposures may not play a role in the pathogenesis of Modic changes on the lumbar spine MR images. Spine J 20(1):94–100. https://doi.org/10.1016/j.spinee.2019.08.009 (PubMed PMID: 31442618)
Braten LCH, Grovle L, Espeland A et al (2020) Clinical effect modifiers of antibiotic treatment in patients with chronic low back pain and Modic changes - secondary analyses of a randomised, placebo-controlled trial (the AIM study). BMC Musculoskelet Disord 21(1):458. https://doi.org/10.1186/s12891-020-03422-y.PubMedPMID:32660517;PubMedCentralPMCID:PMCPMC7359501
Dudli S, Fields AJ, Samartzis D et al (2016) Pathobiology of Modic changes. Eur Spine J 25(11):3723–3734. https://doi.org/10.1007/s00586-016-4459-7.PubMedPMID:26914098;PubMedCentralPMCID:PMCPMC5477843
Bogl LH, Kaye SM, Ramo JT et al (2016) Abdominal obesity and circulating metabolites: A twin study approach. Metabolism 65(3):111–121. https://doi.org/10.1016/j.metabol.2015.10.027 (PubMed PMID: 26892522)
Karjalainen MK, Holmes MV, Wang Q et al (2020) Apolipoprotein A-I concentrations and risk of coronary artery disease: A Mendelian randomization study. Atherosclerosis 299:56–63. https://doi.org/10.1016/j.atherosclerosis.2020.02.002 (PubMed PMID: 32113648)
Wurtz P, Kangas AJ, Soininen P et al (2017) Quantitative Serum Nuclear Magnetic Resonance Metabolomics in Large-Scale Epidemiology: A Primer on -Omic Technologies. Am J Epidemiol 186(9):1084–1096. https://doi.org/10.1093/aje/kwx016.PubMedPMID:29106475;PubMedCentralPMCID:PMCPMC5860146
Zhai G, Wang-Sattler R, Hart DJ, et al. (2010) Serum branched-chain amino acid to histidine ratio: a novel metabolomic biomarker of knee osteoarthritis. Annals of the rheumatic diseases.69(6):1227–31. Epub 2010/04/15. doi: https://doi.org/10.1136/ard.2009.120857. PubMed PMID: 20388742.
Xia J, Mandal R, Sinelnikov IV, et al. (2012) MetaboAnalyst 2.0--a comprehensive server for metabolomic data analysis. Nucleic Acids Res.40(Web Server issue):W127–33. doi: https://doi.org/10.1093/nar/gks374. PubMed PMID: 22553367; PubMed Central PMCID: PMCPMC3394314.
Purcell S, Neale B, Todd-Brown K et al (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81(3):559–575. https://doi.org/10.1086/519795.PubMedPMID:17701901;PubMedCentralPMCID:PMCPMC1950838
Wigginton JE, Cutler DJ, Abecasis GR (2005) A note on exact tests of Hardy-Weinberg equilibrium. Am J Hum Genet 76(5):887–893. https://doi.org/10.1086/429864.PubMedPMID:15789306;PubMedCentralPMCID:PMCPMC1199378
Price AL, Patterson NJ, Plenge RM et al (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38(8):904–909. https://doi.org/10.1038/ng1847 (PubMed PMID: 16862161)
Kettunen J, Demirkan A, Wurtz P et al (2016) Genome-wide study for circulating metabolites identifies 62 loci and reveals novel systemic effects of LPA. Nat Commun 7:11122. https://doi.org/10.1038/ncomms11122.PubMedPMID:27005778;PubMedCentralPMCID:PMCPMC4814583
Mak TSH, Porsch RM, Choi SW et al (2017) Polygenic scores via penalized regression on summary statistics. Genet Epidemiol 41(6):469–480. https://doi.org/10.1002/gepi.22050 (PubMed PMID: 28480976)
Rhee EP, Ho JE, Chen MH et al (2013) A genome-wide association study of the human metabolome in a community-based cohort. Cell Metab 18(1):130–143. https://doi.org/10.1016/j.cmet.2013.06.013.PubMedPMID:23823483;PubMedCentralPMCID:PMCPMC3973158
Gieger C, Geistlinger L, Altmaier E, et al. (2008) Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet.4(11):e1000282. doi: https://doi.org/10.1371/journal.pgen.1000282. PubMed PMID: 19043545; PubMed Central PMCID: PMCPMC2581785 products and services in the field of targeted quantitative metabolomics research. The other authors have no competing interests to declare.
Colhoun HM, Otvos JD, Rubens MB et al (2002) Lipoprotein subclasses and particle sizes and their relationship with coronary artery calcification in men and women with and without type 1 diabetes. Diabetes 51(6):1949–1956. https://doi.org/10.2337/diabetes.51.6.1949 (PubMed PMID: 12031985)
Ala-Korpela M. (2008) Critical evaluation of 1H NMR metabonomics of serum as a methodology for disease risk assessment and diagnostics. Clin Chem Lab Med.46(1):27–42. Epub 2007/11/21. doi: https://doi.org/10.1515/CCLM.2008.006. PubMed PMID: 18020967.
Tukiainen T, Tynkkynen T, Makinen VP, et al. (2008) A multi-metabolite analysis of serum by 1H NMR spectroscopy: early systemic signs of Alzheimer's disease. Biochem Biophys Res Commun.375(3):356–61. Epub 2008/08/14. doi: S0006–291X(08)01520–9 [pii] https://doi.org/10.1016/j.bbrc.2008.08.007. PubMed PMID: 18700135.
Makinen VP, Soininen P, Forsblom C, et al. (2008) 1H NMR metabonomics approach to the disease continuum of diabetic complications and premature death. Mol Syst Biol.4:167. Epub 2008/02/16. doi: msb4100205 [pii] https://doi.org/10.1038/msb4100205. PubMed PMID: 18277383; PubMed Central PMCID: PMC2267737.
Goonewardena SN, Prevette LE, Desai AA. (2010) Metabolomics and atherosclerosis. Curr Atheroscler Rep.12(4):267–72. Epub 2010/05/14. doi: https://doi.org/10.1007/s11883-010-0112-9. PubMed PMID: 20464531.
Inouye M, Kettunen J, Soininen P, et al. (2010) Metabonomic, transcriptomic, and genomic variation of a population cohort. Mol Syst Biol.6:441. Epub 2010/12/24. doi: https://doi.org/10.1038/msb.2010.93. PubMed PMID: 21179014; PubMed Central PMCID: PMC3018170.
Inouye M, Ripatti S, Kettunen J, et al. (2012) Novel Loci for metabolic networks and multi-tissue expression studies reveal genes for atherosclerosis. PLoS Genet.8(8):e1002907. Epub 2012/08/24. doi: https://doi.org/10.1371/journal.pgen.1002907. PubMed PMID: 22916037; PubMed Central PMCID: PMC3420921.
Wurtz P, Soininen P, Kangas AJ et al (2011) Characterization of systemic metabolic phenotypes associated with subclinical atherosclerosis. Mol Biosyst 7(2):385–393. https://doi.org/10.1039/c0mb00066c (PubMed PMID: 21057674)
Acar IE, Lores-Motta L, Colijn JM et al (2020) Integrating metabolomics, genomics, and disease pathways in age-related macular degeneration: the EYE-RISK consortium. Ophthalmology. https://doi.org/10.1016/j.ophtha.2020.06.020 (PubMed PMID: 32553749)
Kauppila LI, McAlindon T, Evans S, et al. (1997) Disc degeneration/back pain and calcification of the abdominal aorta. A 25-year follow-up study in Framingham. Spine.22(14):1642–7; discussion 8–9. PubMed PMID: 9253101.
Acknowledgements
This work was supported by grants from the Hong Kong Theme-Based Research Scheme (T12-708/12N), Hong Kong Research Grants Council (776613), and the Hansjorg Wyss Award by AO Spine International. The authors would like to thank Ms. Cora Bow, Yu Pei, and Kenneth MC Cheung from the Department of Orthopaedics and Traumatology, Hong Kong for their assistance with the cohort. The authors would also like thank Drs. Mika Ala-Korpela, Pasi Soininen and Antti J Kangas from the University of Eastern Finland, NMR Metabolomics Laboratory, Finland, for their assistance in analyzing the blood samples to obtain the metabolomic analytes.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no financial or competing interests related to this work.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Y., Karppinen, J., Cheah, K.S.E. et al. Integrative analysis of metabolomic, genomic, and imaging-based phenotypes identify very-low-density lipoprotein as a potential risk factor for lumbar Modic changes . Eur Spine J 31, 735–745 (2022). https://doi.org/10.1007/s00586-021-06995-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00586-021-06995-x