Abstract
Purpose
To evaluate stability and machine learning-based classification performance of radiomic features of spine bone tumors using diffusion- and T2-weighted magnetic resonance imaging (MRI).
Material and methods
This retrospective study included 101 patients with histology-proven spine bone tumor (22 benign; 38 primary malignant; 41 metastatic). All tumor volumes were manually segmented on morphologic T2-weighted sequences. The same region of interest (ROI) was used to perform radiomic analysis on ADC map. A total of 1702 radiomic features was considered. Feature stability was assessed through small geometrical transformations of the ROIs mimicking multiple manual delineations. Intraclass correlation coefficient (ICC) quantified feature stability. Feature selection consisted of stability-based (ICC > 0.75) and significance-based selections (ranking features by decreasing Mann–Whitney p-value). Class balancing was performed to oversample the minority (i.e., benign) class. Selected features were used to train and test a support vector machine (SVM) to discriminate benign from malignant spine tumors using tenfold cross-validation.
Results
A total of 76.4% radiomic features were stable. The quality metrics for the SVM were evaluated as a function of the number of selected features. The radiomic model with the best performance and the lowest number of features for classifying tumor types included 8 features. The metrics were 78% sensitivity, 68% specificity, 76% accuracy and AUC 0.78.
Conclusion
SVM classifiers based on radiomic features extracted from T2- and diffusion-weighted imaging with ADC map are promising for classification of spine bone tumors. Radiomic features of spine bone tumors show good reproducibility rates.
Similar content being viewed by others
Introduction
Bone tumors of the spine include benign and malignant entities. The incidence of benign lesions, such as hemangioma, is hard to determine as they are often asymptomatic and remain undetected or are discovered incidentally. Among malignant bone tumors of the spine, metastatic lesions are far more common than primary lesions [1]. Imaging, and particularly magnetic resonance imaging (MRI), plays a pivotal role in the discrimination among these entities [2]. Although MRI performed with conventional pulse sequences has good reliability, some features of benign and malignant spinal lesions overlap and make the differential diagnosis challenging [3]. Diffusion-weighted (DW) imaging provides information regarding tumor cellularity. Specifically, apparent diffusion coefficient (ADC) maps are quantitative measures of the magnitude of diffusion within tissues. The role of DW imaging has been highlighted in previous studies dealing with bone tumors of the spine [4], and mean ADC values have been shown to discriminate between benign and malignant entities, including both primary malignant and metastatic lesions [5].
Radiomics includes extraction, analysis and interpretation of large numbers of quantitative data, known as radiomic features, from medical imaging [6, 7]. Radiomics has gained attention in oncologic imaging, and most studies to date have focused on discriminating tumor grades and types before treatment, monitoring response to therapy and predicting outcome [8]. A great variability in radiomic features has, however, emerged as a major issue across these studies, particularly with regard to the segmentation process, thus highlighting the need for preliminary assessment of feature stability [9]. Machine learning aids in analyzing radiomic data, as it performs inferences from large amounts of radiomic features and creates classification models for the diagnosis of interest [10, 11].
The objectives of this study were to evaluate stability and machine learning-based classification performance of radiomic features of spine bone tumors extracted from DW and T2-weighted magnetic resonance imaging (MRI). ADC value differences among benign, primary malignant and metastatic tumors of the spine were also assessed.
Materials and methods
Dataset description
Institutional Review Board approval and a waiver for informed consent were obtained. This retrospective study included 101 patients with histology-proven spine bone tumors and DW MRI performed over the last 10 years at one tertiary bone tumor center. Of the 101 patients used for the study, 22 were benign and 79 were malignant (38 primary and 41 metastatic). Clinical and demographic characteristics are summarized in Table 1.
All images were acquired using 1.5 T MRI scanners and different image acquisition parameters (pixel spacing, slice thickness, time of repetition and echo). T2-weighted and DW images were available for all patients included. T2-weighted images were acquired using turbo spin-echo pulse sequences, while DW images were acquired using echo-planar imaging with b-values of 0 and 1000 s/mm2. Further details on MRI acquisition are displayed in Table 2. The DW images were used to fit ADC maps.
Image segmentation
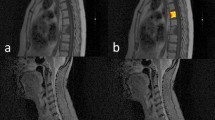
3D image segmentation was manually performed by a last-year resident in radiology experienced in musculoskeletal and oncologic imaging using the open-source software ITK-SNAP (version 3.6) [12]. The reader drew a region of interest (ROI) along tumor borders on each slice using axial T2-weighted sequence, which provided more morphological details compared to DW images. Since there was not an excessive movement of the patient from one sequence to the other, the same segmentation was used as a mask to extract the radiomic features from both ADC and T2-weighted images. An example of segmentation overlaid on both T2-weighted and ADC images is displayed in Fig. 1.
Image preprocessing
Different preprocessing steps were applied to the MRI images prior to the extraction of the radiomic features. First, a 3D Gaussian filter with a 3 × 3× 3 voxel kernel and σ = 0.5 was used to denoise the images. In case of T2-weighted images, a bias field correction was performed using the N4ITK algorithm [13] to correct for potential effects due to inhomogeneity of the magnetic field. Also, Z-score standardization was performed to ensure that the range of intensity in the T2-weighted images was the same. Last, the images were resampled to a common isotropic resolution of 2 mm (as in [14]) using B-spline interpolation. Bias field correction and intensity standardization were not applied on the ADC maps because those types of images already have a standardized range of intensity (diffusion coefficients are within the maximum range of 0–4 10–3 mm2/s) and are not affected magnetic field inhomogeneity.
Radiomic features extraction
An initial set of 1702 radiomic features was extracted from the MRI volumes (851 features per sequence) using the PyRadiomics library (version 3.0) [15]. Features belonged to different categories such as shape and size, first-order statistics (FOS) and textural features. Textural features were computed using five different textural matrices: the gray-level co-occurrence matrix (GLCM); the gray-level run length matrix (GLRLM); the gray-level size zone matrix (GLSZM); the neighboring gray tone difference matrix (NGTDM); and the gray-level dependence matrix (GLDM). FOS and textural features were extracted from both the original volumes and the 8 volumes obtained by first-level wavelet decomposition [16]. For the full list of radiomic features, refer to PyRadiomics documentation (https://pyradiomics.readthedocs.io/en/latest/features.html) and to supplementary material “Supplementary 1 – Features description.” A 32-bins gray-level discretization was used prior to the radiomic features extraction.
Stability analysis
Prior to the training of any radiomic model, a side experiment was performed to evaluate the stability of radiomic features to variations in the ROI. This stability analysis was performed to mimic the effect of potential sources of variability such as the intra- and inter-reader variability that comes with manual segmentation or the potential mismatch between the ROI and the underlying tumor when the same ROI is used to extract features from multiple sequences; like it was done in this study. The stability analysis was performed as described in [17], by applying different translation of the ROI in the positive and negative direction of the x and y directions, respectively (Fig. 2). The entity of the translation was 10% of the length of the bounding box of the tumor in the corresponding direction (x or y). For each patient, the radiomic features were extracted from 5 different ROIs (the original and the 4 translated versions). Intraclass correlation coefficient (ICC) was used to quantify the stability of each radiomic feature [18]. A feature was considered stable if ICC was higher than 0.75 [19].
Radiomic classifier training and validation pipeline
Figure 3 shows the scheme including all the steps for the training and validation of the radiomics-based classifier. First, according to the previous stability analysis, radiomic features having an ICC of 0.75 or lower were consider unstable and excluded. The second step of features selection was related to the ability of radiomic features to discriminate between malignant and benign tumors. Mann–Whitney tests were used to identify features with a statistically different distribution between the two groups. The statistically different features were kept and ranked by their p-value (from lower to higher). Of these ranked features, only the first n were selected (with n ranging from 1 to 10).
Prior to the training of the classifier, a class balancing was applied using the Synthetic Minority Oversampling Technique (SMOTE) [20], which is a technique used to artificially oversample the minority class (in this case, the benign tumors). The balanced dataset was used to train a support vector machine (SVM) classifier to discriminate the type of tumor. The reference “positive” group for the training of the classifier was the group of malignant tumors. The training was performed in MATLAB using fitcsvm function with the default hyperparameters (see Name-Value Pair Arguments in https://it.mathworks.com/help/stats/fitcsvm.html) and a linear kernel.
Radiomic classifier validation
The diagnostic performance of the radiomic model was evaluated using tenfold cross-validation on the training set. In each iteration, the training pipeline was applied to 9/10 of the dataset and the train model was used to classify the remaining patients. For each patient, an unbiasedly evaluated class and a classification score could be computed. Using these two elements, both a confusion matrix and a receiver operating characteristics (ROC) curve could be computed. Sensitivity, specificity and accuracy of the classification as well as the area under the ROC curve (AUC) were determined and used to evaluate the quality of the classifier. In the computation of these metrics, the group of malignant tumors was considered as the “positive” class. The number of selected features used to train the different model was varied from 1 to 10 to evaluate the effect of an increasing number of features on model’s performance.
Discriminative power of mean ADC
Since previous studies [5] reported the ability of mean ADC in discriminating the different categories of tumor (benign, primary malignant or metastatic), we tried to confirm this ability, using the mean ADC obtained by the ROI of this dataset. A Kruskal–Wallis test and post hoc comparisons were performed to evaluate whether mean ADC had a significantly different distribution among the groups. Also, the AUC of mean ADC for the benign vs. malignant classification was evaluated.
Results
Stability analysis
A total of 1300 extracted radiomic features (76.4%) were stable to transformations of the ROI. The full list of radiomic features used for the following analysis is detailed in supplementary material “Supplementary 2—Features stability.xlsx.”
Radiomic classifier validation
The quality metrics for the SVM models are displayed in Table 3 as a function of the number of selected features. The model with the best performance and the lowest number of features was the model with 8 features, with a sensitivity of 0.78, specificity of 0.68 and accuracy of 0.76 (Fig. 4a). The ROC curve of the best model is displayed in Fig. 4b. Its area under the ROC curve (AUC) was 0.78.
Discriminative power of mean ADC
The boxplots in Fig. 5a show the distribution of mean ADC among the different groups of tumors (benign, primary malignant and metastasis). The values of mean ADC were 1.30 ± 0.35 *10–3 mm2/s for benign tumors, 1.17 ± 0.24 *10–3 mm2/s for metastasis and 1.08 ± 0.36 *10–3 mm2/s for primary malignant tumors. The p-value for the Kruskal–Wallis test was 0.02, and post hoc comparisons showed that the difference between benign and primary malignant tumors was statistically significant (p = 0.01) but not between benign tumors and metastasis (p = 0.38). The AUC of mean ADC for benign vs. malignant tumor was 0.66.
Boxplots showing the distribution of the values of mean apparent diffusion coefficient (ADC). (a) Values of this study. (b) Distributions observed in [5]
Discussion
The main finding of this study is that SVM models based on radiomic features derived from T2-weighted and DW MRI with ADC map allowed for benign vs. malignant classification of spine bone tumors with up to 76% accuracy. Additionally, the reproducibility rate of radiomic features was higher than 76% as features were stable to geometrical transformations of the ROIs.
Previous studies dealt with MRI radiomics-based classification of spine bone tumors. In cancer patients with bone marrow metastatic disease, a very preliminary investigation showed that MRI-based radiomic signature could be helpful to discriminate between metastatic and non-metastatic vertebral bodies [21]. Recently, Chianca et al. tested different extraction software for radiomics-based classification of spine bone tumors using T1-weighted and T2-weighted MRI and reported that PyRadiomics outperformed other extraction software [22]. The reason is likely that PyRadiomics directly enables robust image preprocessing, thus removing dependencies on some image parameters and allowing for generalizability of the results, as it was done in our study. Lang et al. [23] focused on dynamic contrast-enhanced MRI and differentiated spine metastatic lesions originated from lung and other cancers using radiomics and deep learning. In a dataset of 30 metastases from lung cancer and 31 metastases from other cancers, classification using convolutional neural networks achieved a mean accuracy of 71% [23]. Our current study adds to the literature by highlighting the role of machine-learning classification of spine bone tumors based on radiomic features extracted from DW MRI with ADC maps, coupled with morphologic T2-weighted sequences. A SVM was trained and tested based on selected radiomic features and had an accuracy of up to 76%.
Stability analysis allows for assessing the robustness of radiomic features and represents a preliminary step in the process of feature selection. Several strategies can be used for stability evaluation, such as changes in image acquisition parameters [24] and multiple ROI delineations performed by different readers [25], which are, however, time-consuming. In our study, feature stability was assessed through a time-saving method based on geometrical transformations of the ROIs mimicking multiple manual delineations [17]. More than 76% radiomic features were stable to these transformations and then showed good overall reproducibility.
A recent meta-analysis showed that quantitative assessment of ADC was excellent for differentiating benign from malignant bone marrow lesions of the spine [4]. In a study included in this meta-analysis, mean ADC values were found to be higher in benign bone tumors in comparison with both primary malignant and metastatic lesions of the spine [5]. Our results are in line with those findings (Fig. 5B), as benign tumors showed higher ADC values than primary malignant lesions and metastases, although statistical significance was not reached in the latter case. The AUC of mean ADC for benign vs. malignant tumor discrimination was 0.66 and 0.76 in our and previous [5] studies, respectively. However, an overlap between mean ADC values of benign and malignant tumors was highlighted in the previous investigation, e.g., giant cell tumor of the bone is histologically benign but has low ADC mean value [5]. In this regard, an objective assessment of tumor heterogeneity by means of radiomics and machine learning might aid in the diagnosis.
Some limitations of this study need to be taken into account. First, it has a retrospective design as a prospective analysis is not strictly necessary for radiomic studies [8]. Second, the number of histology-proven benign lesions was small, i.e., approximately one fifth of the population of study, because histology was not available for unbiopsied benign lesions with typical imaging findings. However, this limitation was overcome by means of class balancing with SMOTE technique [20]. Finally, an independent cohort of patients was not available for external validation of the radiomics-based classifier and needs to be included in further investigations.
Limitations notwithstanding, we conclude that SVM models based on radiomic features extracted from T2-weighted and DW MRI with ADC map are promising for classification of bone tumors of the spine and radiomic features show good overall reproducibility.
Availability of data and material
Data may be obtained from the corresponding author on reasonable request.
Code availability
Data may be obtained from the corresponding author on reasonable request.
Abbreviations
- ADC:
-
Apparent diffusion coefficient
- AUC:
-
Area under the ROC curve
- DW:
-
Diffusion weighted
- FOS:
-
First-order statistics
- GLCM:
-
Gray-level co-occurrence matrix
- GLDM:
-
Gray-level dependence matrix
- GLRLM:
-
Gray-level run length matrix
- GLSZM:
-
Gray-level size zone matrix
- ICC:
-
Intraclass correlation coefficient
- MRI:
-
Magnetic resonance imaging
- NGTDM:
-
Neighboring gray tone difference matrix
- ROC:
-
Receiver operating characteristics
- ROI:
-
Region of interest
- SMOTE:
-
Synthetic minority oversampling technique
- SVM:
-
Support vector machine
References
Albano D, Messina C, Gitto S et al (2019) Differential diagnosis of spine tumors: my favorite mistake. Semin Musculoskelet Radiol 23:026–035. https://doi.org/10.1055/s-0038-1675551
Cannavò L, Albano D, Messina C et al (2019) Accuracy of CT and MRI to assess resection margins in primary malignant bone tumours having histology as the reference standard. Clin Radiol 74:736.e13-736.e21. https://doi.org/10.1016/j.crad.2019.05.022
Jung H-S, Jee W-H, McCauley TR et al (2003) Discrimination of metastatic from acute osteoporotic compression spinal fractures with MR imaging 1. Radiographics 23:179–187. https://doi.org/10.1148/rg.231025043
Suh CH, Yun SJ, Jin W et al (2018) ADC as a useful diagnostic tool for differentiating benign and malignant vertebral bone marrow lesions and compression fractures: a systematic review and meta-analysis. Eur Radiol 28:2890–2902. https://doi.org/10.1007/s00330-018-5330-5
Pozzi G, Albano D, Messina C et al (2018) Solid bone tumors of the spine: Diagnostic performance of apparent diffusion coefficient measured using diffusion-weighted MRI using histology as a reference standard. J Magn Reson Imaging 47:1034–1042. https://doi.org/10.1002/jmri.25826
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577. https://doi.org/10.1148/radiol.2015151169
Gitto S, Cuocolo R, Albano D et al (2021) CT and MRI radiomics of bone and soft-tissue sarcomas: a systematic review of reproducibility and validation strategies. Insights Imaging 12:68. https://doi.org/10.1186/s13244-021-01008-3
Lubner MG, Smith AD, Sandrasegaran K et al (2017) CT texture analysis: definitions, applications, biologic correlates, and challenges. Radiographics 37:1483–1503. https://doi.org/10.1148/rg.2017170056
Gitto S, Cuocolo R, Emili I et al (2021) Effects of interobserver variability on 2D and 3D CT- and MRI-based texture feature reproducibility of cartilaginous bone tumors. J Digit Imaging. https://doi.org/10.1007/s10278-021-00498-3
Gitto S, Cuocolo R, Albano D et al (2020) MRI radiomics-based machine-learning classification of bone chondrosarcoma. Eur J Radiol 128:109043. https://doi.org/10.1016/j.ejrad.2020.109043
Gitto S, Cuocolo R, Annovazzi A et al (2021) CT radiomics-based machine learning classification of atypical cartilaginous tumours and appendicular chondrosarcomas. EBioMedicine 68:103407. https://doi.org/10.1016/j.ebiom.2021.103407
Yushkevich PA, Piven J, Hazlett HC et al (2006) User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 31:1116–1128. https://doi.org/10.1016/j.neuroimage.2006.01.015
Tustison NJ, Cook PA, Gee JC (2010) N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 29:1310–1320. https://doi.org/10.1109/TMI.2010.2046908.N4ITK
Leijenaar RT, Bogowicz M, Jochems A et al (2018) Development and validation of a radiomic signature to predict HPV (p16) status from standard CT imaging: a multicenter study. Br J Radiol 91:20170498. https://doi.org/10.1259/bjr.20170498
van Griethuysen JJM, Fedorov A, Parmar C et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77:e104–e107. https://doi.org/10.1158/0008-5472.CAN-17-0339
Aerts HJWL, Velazquez ER, Leijenaar RTH et al (2014) Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 5:4006
Bologna M, Corino VDA, Montin E et al (2018) Assessment of stability and discrimination capacity of radiomic features on apparent diffusion coefficient images. J Digit Imaging 31:879–894. https://doi.org/10.1007/s10278-018-0092-9
Mcgraw KO (1996) Forming inferences about some intraclass correlation coefficients. Psychol Methods 1:30–46. https://doi.org/10.1037/1082-989X.1.1.30
Koo TK, Li MY (2016) A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med 15:155–163. https://doi.org/10.1016/j.jcm.2016.02.012
Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP (2002) SMOTE: synthetic minority over-sampling technique. J Artif Intell Res 16:321–357. https://doi.org/10.1613/jair.953
Filograna L, Lenkowicz J, Cellini F et al (2019) Identification of the most significant magnetic resonance imaging (MRI) radiomic features in oncological patients with vertebral bone marrow metastatic disease: a feasibility study. Radiol Med 124:50–57. https://doi.org/10.1007/s11547-018-0935-y
Chianca V, Cuocolo R, Gitto S et al (2021) Radiomic machine learning classifiers in spine bone tumors: a multi-software. Multi-Scanner Study Eur J Radiol 137:109586. https://doi.org/10.1016/j.ejrad.2021.109586
Lang N, Zhang Y, Zhang E et al (2019) Differentiation of spinal metastases originated from lung and other cancers using radiomics and deep learning based on DCE-MRI. Magn Reson Imaging 64:4–12. https://doi.org/10.1016/j.mri.2019.02.013
Galavis PE, Hollensen C, Jallow N et al (2010) Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters. Acta Oncol (Madr) 49:1012–1016. https://doi.org/10.3109/0284186X.2010.498437
Gitto S, Cuocolo R, van Langevelde K et al (2022) MRI radiomics-based machine learning classification of a typical cartilaginous tumour and grade II chondrosarcoma of long bones. EBioMedicine 75:103757. https://doi.org/10.1016/j.ebiom.2021.103757
Funding
Open access funding provided by Università degli Studi di Milano within the CRUI-CARE Agreement.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
Institutional Review Board approval was obtained, and the need for written informed consent was waived. All procedures involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gitto, S., Bologna, M., Corino, V.D.A. et al. Diffusion-weighted MRI radiomics of spine bone tumors: feature stability and machine learning-based classification performance. Radiol med 127, 518–525 (2022). https://doi.org/10.1007/s11547-022-01468-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11547-022-01468-7